Vibrational spectroscopy#
Vibrational spectroscopy techniques are widely used in chemistry for the identification of compounds or functional groups in the three states of matter: solid, liquid and gaseous state. In this chapter, we are going to revisit and ilustrate some basic concepts to uderstand the physical phenomena behind the main techniques, which are infrared spectroscopy and Raman spectroscopy.
Rayleigh scattering#
History
In 1871 Lord Rayleigh explains the colour and polarisation of the skylight. Rayleigh scattering is an elastic scattering of electromagnetic radiation by particles. The oscillating electric field of light excites the charges of a molecule, causing them to vibrate at the same frequency. The molecule becomes a small radiating dipole scattering the incoming light.
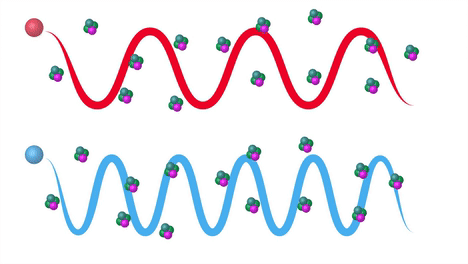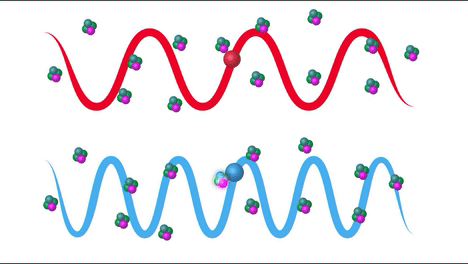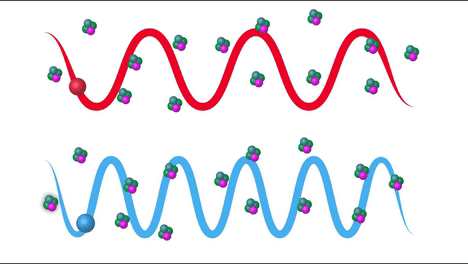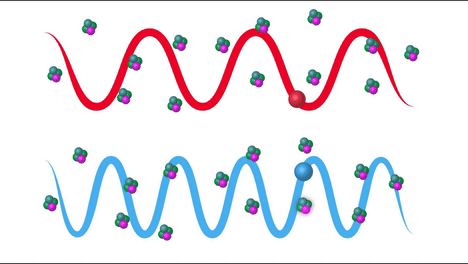
Raman Effect#
History
Sir C. V. Raman and K. S. Krishnan observed the effect in organic liquids using sunlight in 1928, Raman obtained the Nobel prize in physics in 1930

Fig. 1 C.V. Raman with his spectrometer#
Raman scattering or the Raman effect is the inelastic scattering of photons by matter, meaning that there is both an exchange of energy and a change in the light’s direction. Typically this effect involves vibrational energy being gained by a molecule as incident photons from a visible laser are shifted to lower energy. This is called normal Stokes Raman scattering. If the incident photons are shifted to higher energy the process is called Anti-Stokes Raman scattering
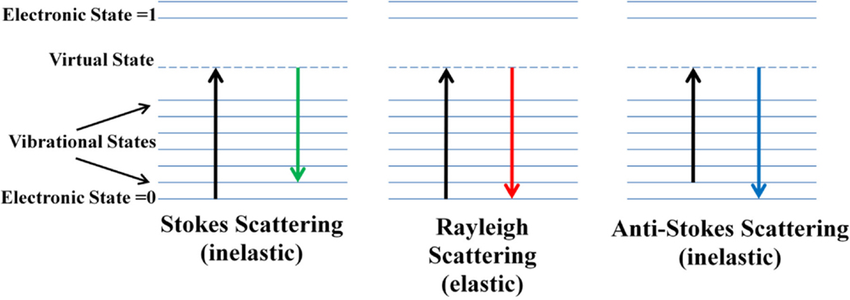
Here we can see how the incident beam produces diferent frequencies depending on the type of scattering:
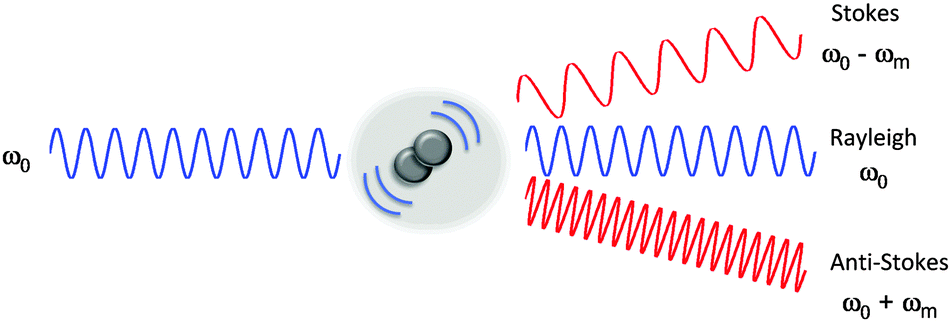
Spectrum of light#
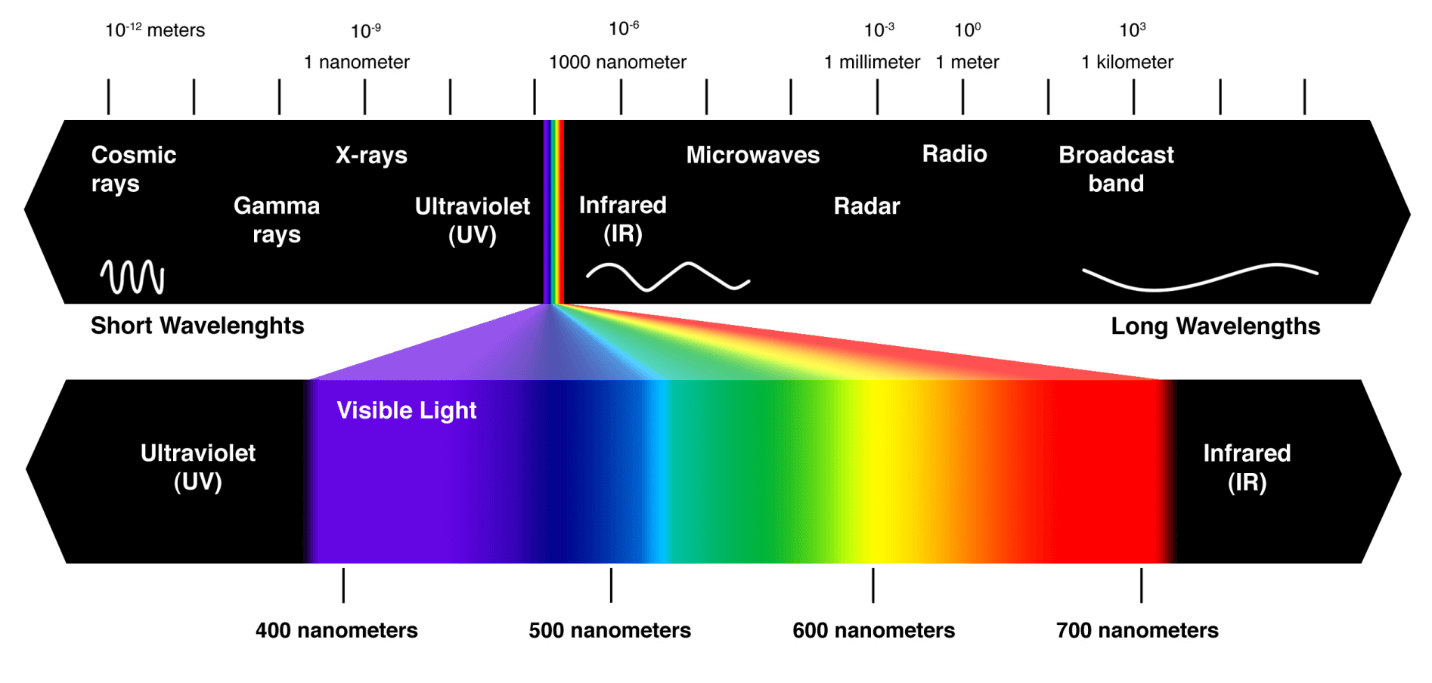
Spectroscopy techniques are available over a very wide energy range:
Gamma rays: Mössbauer spectroscopy
X-rays: X-ray photoelectron spectroscopy
UV-Vis: Raman spectroscopy
IR: IR spectroscopy
Microwave: Electron paramagnetic resonance spectroscopy
Radiowave: Nuclear magnetic resonance
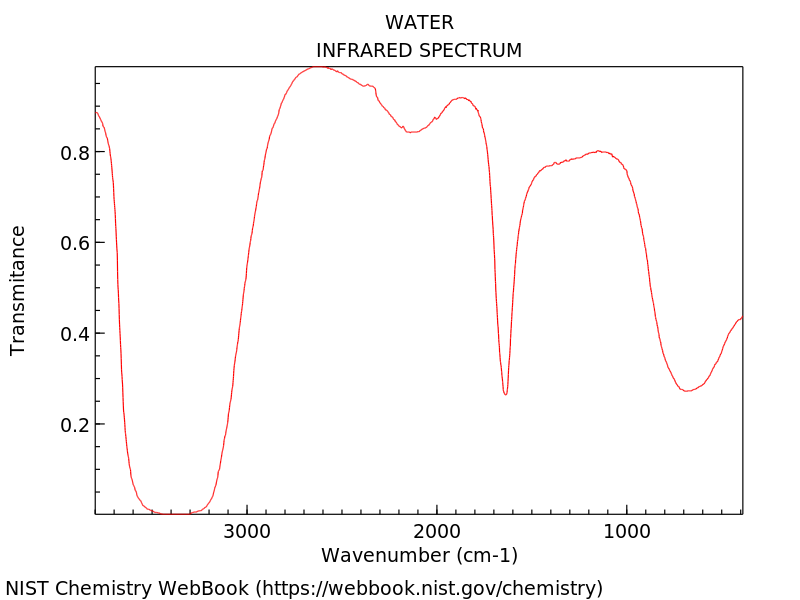
Infrared Spectroscopy#
Infrared spectroscopy (IR spectroscopy) is the measurement of the interaction of infrared radiation with matter by absorption, emission, or reflection. It is used to study and identify chemical substances or functional groups. The identifications proceeds via obtaining the infrared spectrum. An IR spectrum can be visualized in a graph of infrared light absorbance (or transmittance) on the vertical axis vs the wavenumber. The wave number is a unit directric proportional to the energy.
Here we show the IR spectrum of the water, in a plot of trasmitance vs wavenumber:
Clasical approach#
We can start a first approach from a clasical point of view. Imagine that we have two atoms joined by a spring, we can actually used Hook Law to derive the force that each atom experiences:
The armonic potential \(V(R)\) can be expanded in Taylor series around the equilibrium position \(R_0\):
Being the thrid term the spring constant \(k\)
Euler equation approach#
import matplotlib.pyplot as plt
import imageio
from pylab import *
---------------------------------------------------------------------------
ModuleNotFoundError Traceback (most recent call last)
Cell In[1], line 2
1 import matplotlib.pyplot as plt
----> 2 import imageio
3 from pylab import *
ModuleNotFoundError: No module named 'imageio'
e=1 # electron charge
s=1 # interatomic distance
m1=10 #mass 1
m1_b=1500
m2=10 #mass 2
k=10 #string constant
k2=150 #string constant
m=m1*m2/(m1+m2) #reduced mass
def vibra(part,p_0,k, m1, m2):
FV=zeros((part,2))
bond=2.*s
m=m1*m2/(m1+m2)
for at in range(0,part,2):
d= p_0[at]-p_0[at+1] #vector distancia enlace entre un particula y su consecutiva
r= sqrt(d[0]**2+d[1]**2) # norma del vector
u=d/r
FV[at]=-k*(r-bond)*u
FV[at+1]=-k*(-r+bond)*u
acelv=FV/m
return acelv
def graf(pn_0,vc_0,pn_k,vc_k,pn_m,vc_m):
alpha_1=0.25
p_0=array([[0,2], [1, 2]]) # positions
p_k=array([[0,0], [1, 0]]) # positions
p_m=array([[0,4], [1, 4]]) # positions
ax.clear() #borra la graficar y pone otra encima
ax.set_xlim(-3,4) #ax tiene ese atributo porque se definió abajo
ax.set_ylim(-3,box)
ax.set_xlabel("x")
ax.set_ylabel("y")
#Change in m
m1,m2= 10,20
x1_m=pn_m[:,0][0]
y1_m=pn_m[:,1][0]
ax.scatter(x1_m,y1_m,s=m1*50, c="blue",edgecolor="None")
x2_m=pn_m[:,0][1]
y2_m=pn_m[:,1][1]
ax.scatter(x2_m,y2_m,s=m2*50,c="orange",edgecolor="None")
ax.hlines(y2_m, xmin=x1_m, xmax=x2_m, linewidth=2, color='green', label="Variation in m")
ax.scatter(p_m[:,0][0],p_m[:,1][0],s=m1*50, c="blue",edgecolor="None", alpha=alpha_1)
ax.scatter(p_m[:,0][1],p_m[:,1][1],s=m2*50, c="blue",edgecolor="None", alpha=alpha_1)
### Reference
m1,m2= 10,10
x1_0=pn_0[:,0][0]
y1_0=pn_0[:,1][0]
ax.scatter(x1_0,y1_0,s=m1*50, c="blue",edgecolor="None")
x2_0=pn_0[:,0][1]
y2_0=pn_0[:,1][1]
ax.scatter(x2_0,y2_0,s=m2*50,c="orange",edgecolor="None")
ax.hlines(y2_0, xmin=x1_0, xmax=x2_0, linewidth=2, color='black', label="Reference")
ax.scatter(p_0[:,0][0],p_0[:,1][0],s=m1*50, c="blue",edgecolor="None", alpha=alpha_1)
ax.scatter(p_0[:,0][1],p_0[:,1][1],s=m1*50, c="blue",edgecolor="None", alpha=alpha_1)
#Change in k
x1_k=pn_k[:,0][0]
y1_k=pn_k[:,1][0]
ax.scatter(x1_k,y1_k,s=m1*50, c="blue",edgecolor="None")
x2_k=pn_k[:,0][1]
y2_k=pn_k[:,1][1]
ax.scatter(x2_k,y2_k,s=m2*50,c="orange",edgecolor="None")
ax.hlines(y2_k, xmin=x1_k, xmax=x2_k, linewidth=2, color='r', label="Variation in k")
ax.scatter(p_k[:,0][0],p_k[:,1][0],s=m1*50, c="blue",edgecolor="None", alpha=alpha_1)
ax.scatter(p_k[:,0][1],p_k[:,1][1],s=m1*50, c="blue",edgecolor="None", alpha=alpha_1)
leg = plt.legend(loc='lower center')
fig.canvas.draw() #muestra la figura
plt.show()
# #Definir parametros
%matplotlib notebook
box = 5.0
part = 2
# Initial conditions
v_0=array([[0, 0], [1, 0]]) # velocities between -1 y 1
v_k=array([[0, 0], [1, 0]]) # velocities between -1 y 1
v_m=array([[0, 0], [1, 0]]) # velocities between -1 y 1
p_0=array([[0,2], [1, 2]]) # positions
p_k=array([[0,0], [1, 0]]) # positions
p_m=array([[0,4], [1, 4]]) # positions
# #Building the figure
fig, ax = plt.subplots(1, 1,figsize=(6, 5))
# fig=plt.figure()
# ax=fig.add_subplot(111)
# fig.canvas.draw() #update the figure
dt=0.01
tiempo=0
#Start simulation
cont=0
position_0=[]
position_m=[]
position_k=[]
t=[]
while tiempo < 10:
aV_0 = vibra(part,p_0,k,m1,m2)
aV_k = vibra(part,p_k,k2,m1,m2)
aV_m = vibra(part,p_m,k,m1_b,m2)
vn_0 = v_0+aV_0*dt
vn_k = v_k+aV_k*dt
vn_m = v_m+aV_m*dt
#adjust velocities
vn_0[(vn_0>0.5)] =0.5
vn_0[(vn_0<-0.5)]=-0.5
vn_k[(vn_k>0.5)] =0.5
vn_k[(vn_k<-0.5)]=-0.5
vn_m[(vn_m>0.5)] =0.5
vn_m[(vn_m<-0.5)]=-0.5
pn_0=p_0+vn_0*dt
pn_k=p_k+vn_k*dt
pn_m=p_m+vn_m*dt
vc_0=sqrt(vn_0[:,0]**2+vn_0[:,1]**2)
vc_k=sqrt(vn_k[:,0]**2+vn_k[:,1]**2)
vc_m=sqrt(vn_m[:,0]**2+vn_m[:,1]**2)
cont+=1
if cont%10==0:
graf(pn_0,vc_0,pn_k,vc_k,pn_m,vc_m)
tiempo=tiempo+dt
p_0=pn_0
v_0=vn_0
p_k=pn_k
v_k=vn_k
p_m=pn_m
v_m=vn_m
position_0.append(pn_0[:,0][0])
position_m.append(pn_m[:,0][0])
position_k.append(pn_k[:,0][0])
t.append(tiempo)
Coupled systems#
import matplotlib.pyplot as plt
import imageio
from pylab import *
s=1
def vibra(part,p_0,k, m1, m2):
FV=zeros((part,2))
bond=2.*s
m=m1*m2/(m1+m2)
for at in range(0,part,2):
d= p_0[at]-p_0[at+1] #vector distancia enlace entre un particula y su consecutiva
r= sqrt(d[0]**2+d[1]**2) # norma del vector
u=d/r
FV[at]=-k*(r-bond)*u
FV[at+1]=-k*(-r+bond)*u
acelv=FV/m
return acelv
def graf(pn_0,vc_0,pn_k,vc_k,pn_m,vc_m):
alpha_1=0.25
p_0=array([[0,2], [1, 2]]) # positions
p_k=array([[0,0], [1, 0]]) # positions
p_m=array([[0,4], [1, 4]]) # positions
ax[0].clear() #borra la graficar y pone otra encima
ax[0].set_xlim(-3,4) #ax tiene ese atributo porque se definió abajo
ax[0].set_ylim(-3,box)
ax[0].set_xlabel("x")
ax[0].set_ylabel("y")
#Change in m
m1,m2= 10,20
x1_m=pn_m[:,0][0]
y1_m=pn_m[:,1][0]
ax[0].scatter(x1_m,y1_m,s=m1*50, c="blue",edgecolor="None")
x2_m=pn_m[:,0][1]
y2_m=pn_m[:,1][1]
ax[0].scatter(x2_m,y2_m,s=m2*50,c="orange",edgecolor="None")
ax[0].hlines(y2_m, xmin=x1_m, xmax=x2_m, linewidth=2, color='green', label="Variation in m")
ax[0].scatter(p_m[:,0][0],p_m[:,1][0],s=m1*50, c="blue",edgecolor="None", alpha=alpha_1)
ax[0].scatter(p_m[:,0][1],p_m[:,1][1],s=m2*50, c="blue",edgecolor="None", alpha=alpha_1)
### Reference
m1,m2= 10,10
x1_0=pn_0[:,0][0]
y1_0=pn_0[:,1][0]
ax[0].scatter(x1_0,y1_0,s=m1*50, c="blue",edgecolor="None")
x2_0=pn_0[:,0][1]
y2_0=pn_0[:,1][1]
ax[0].scatter(x2_0,y2_0,s=m2*50,c="orange",edgecolor="None")
ax[0].hlines(y2_0, xmin=x1_0, xmax=x2_0, linewidth=2, color='black', label="Reference")
ax[0].scatter(p_0[:,0][0],p_0[:,1][0],s=m1*50, c="blue",edgecolor="None", alpha=alpha_1)
ax[0].scatter(p_0[:,0][1],p_0[:,1][1],s=m1*50, c="blue",edgecolor="None", alpha=alpha_1)
#Change in k
x1_k=pn_k[:,0][0]
y1_k=pn_k[:,1][0]
ax[0].scatter(x1_k,y1_k,s=m1*50, c="blue",edgecolor="None")
x2_k=pn_k[:,0][1]
y2_k=pn_k[:,1][1]
ax[0].scatter(x2_k,y2_k,s=m2*50,c="orange",edgecolor="None")
ax[0].hlines(y2_k, xmin=x1_k, xmax=x2_k, linewidth=2, color='r', label="Variation in k")
ax[0].scatter(p_k[:,0][0],p_k[:,1][0],s=m1*50, c="blue",edgecolor="None", alpha=alpha_1)
ax[0].scatter(p_k[:,0][1],p_k[:,1][1],s=m1*50, c="blue",edgecolor="None", alpha=alpha_1)
ax[0].legend(loc='lower center')
ax[1].clear()
ax[1].set_xlim(0,10) #ax tiene ese atributo porque se definió abajo
ax[1].set_ylim(-1,0)
ax[1].set_xlabel("time")
ax[1].set_ylabel("x")
ax[1].plot(t,position_0, label="Reference",c="black")
ax[1].plot(t,position_m, label="m",c="green")
ax[1].plot(t,position_k, label="k",c="red")
ax[1].legend()
fig.canvas.draw() #muestra la figura
plt.show()
# #Definir parametros
%matplotlib notebook
box = 5.0
part = 2
# Initial conditions
v_0=array([[0, 0], [1, 0]]) # velocities between -1 y 1
v_k=array([[0, 0], [1, 0]]) # velocities between -1 y 1
v_m=array([[0, 0], [1, 0]]) # velocities between -1 y 1
p_0=array([[0,2], [1, 2]]) # positions
p_k=array([[0,0], [1, 0]]) # positions
p_m=array([[0,4], [1, 4]]) # positions
# #Building the figure
fig, ax = plt.subplots(1, 2,figsize=(12, 5))
# fig=plt.figure()
# ax=fig.add_subplot(111)
# fig.canvas.draw() #update the figure
dt=0.01
tiempo=0
#Start simulation
cont=0
position_0=[]
position_m=[]
position_k=[]
t=[]
while tiempo < 10:
aV_0 = vibra(part,p_0,10,10,10)
aV_k = vibra(part,p_k,150,10,10)
aV_m = vibra(part,p_m,10,150,10)
vn_0 = v_0+aV_0*dt
vn_k = v_k+aV_k*dt
vn_m = v_m+aV_m*dt
#adjust velocities
vn_0[(vn_0>0.5)] =0.5
vn_0[(vn_0<-0.5)]=-0.5
vn_k[(vn_k>0.5)] =0.5
vn_k[(vn_k<-0.5)]=-0.5
vn_m[(vn_m>0.5)] =0.5
vn_m[(vn_m<-0.5)]=-0.5
pn_0=p_0+vn_0*dt
pn_k=p_k+vn_k*dt
pn_m=p_m+vn_m*dt
vc_0=sqrt(vn_0[:,0]**2+vn_0[:,1]**2)
vc_k=sqrt(vn_k[:,0]**2+vn_k[:,1]**2)
vc_m=sqrt(vn_m[:,0]**2+vn_m[:,1]**2)
cont+=1
if cont%10==0:
graf(pn_0,vc_0,pn_k,vc_k,pn_m,vc_m)
tiempo=tiempo+dt
p_0=pn_0
v_0=vn_0
p_k=pn_k
v_k=vn_k
p_m=pn_m
v_m=vn_m
position_0.append(pn_0[:,0][0])
position_m.append(pn_m[:,0][0])
position_k.append(pn_k[:,0][0])
t.append(tiempo)
%matplotlib inline
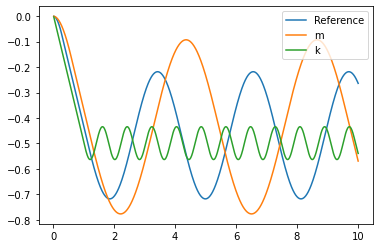
plot(t,position_0, label="Reference")
plot(t,position_m, label="m")
plot(t,position_k, label="k")
leg = plt.legend()
import ipywidgets as widgets
widgets.RadioButtons(
options=['pepperoni', 'pineapple', 'anchovies'],
# value='pineapple', # Defaults to 'pineapple'
# layout={'width': 'max-content'}, # If the items' names are long
description='Pizza topping:',
disabled=False
)
import plotly.express as px
data = px.data.iris()
data.head()
| sepal_length | sepal_width | petal_length | petal_width | species | species_id | |
|---|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa | 1 |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa | 1 |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa | 1 |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa | 1 |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa | 1 |
import plotly.express as px
df = px.data.gapminder()
fig = px.scatter(df, x="gdpPercap", y="lifeExp", animation_frame="year", animation_group="country",
size="pop", color="continent", hover_name="country", facet_col="continent",
log_x=True, size_max=45, range_x=[100,100000], range_y=[25,90])
fig.show()
4961.66/256.75
19.324868549172347
from pylab import *
from ipywidgets import *
@interact(dt=(0,10,0.1), k=(1,100,5), m1=(1,100,10), m2=(1,100,10))
def Vibration(dt=0.1, k=5, m1=10, m2=10):
s=1
box = 5.0
part = 2
# Initial conditions
v_0=array([[0, 0], [1, 0]]) # velocities between -1 y 1
p_0=array([[0, 0], [1, 0]]) # positions
m=m1*m2/(m1+m2) #reduced mass
#Start simulation
def vibra(part,p_0):
FV=zeros((part,2))
bond=2.*s
for at in range(0,part,2):
d= p_0[at]-p_0[at+1] #vector distancia enlace entre un particula y su consecutiva
r= sqrt(d[0]**2+d[1]**2) # norma del vector
u=d/r
FV[at]=-k*(r-bond)*u
FV[at+1]=-k*(-r+bond)*u
acelv=FV/m
return acelv
cont=0
position1=[]
position2=[]
aV=vibra(part,p_0)
aC=aV
vn=v_0+aC*dt
#adjust velocities
vn[(vn>0.5)]=0.5
vn[(vn<-0.5)]=-0.5
pn=p_0+vn*dt
vc=sqrt(vn[:,0]**2+vn[:,1]**2)
p_0=pn
v_0=vn
position1.append(pn[:,0][0])
position2.append(pn[:,0][1])
plt.xlim(-box,box) #ax tiene ese atributo porque se definió abajo
plt.ylim(-box,box)
# ax.set_xlabel("x")
# ax.set_ylabel("y")
x1=pn[:,0][0]
y1=pn[:,1][0]
tiempo=linspace(0,10,len(position1))
a=scatter(tiempo,position1,s=100,c="black",edgecolor="None")
b=scatter(x1,y1,s=m1*50, c="blue",edgecolor="None")
x2=pn[:,0][1]
y2=pn[:,1][1]
c=scatter(x2,y2,s=m2*50,c="orange",edgecolor="None")


